Virus resistant bees?
In the early/mid noughties there was a lot of excitement about a newly discovered pathogen of honey bees, Israeli Acute Paralysis Virus (IAPV). This virus was identified and initially characterised in 2004 and, a couple of years later, was implicated as the (or at least a) potential cause of Colony Collapse Disorder (CCD) {{1}}..
CCD is, and remains (if it still exists at all), enigmatic {{2}}. It is an oft-misused term to describe the dramatic and terminal reduction in worker bee numbers in a colony in the absence of queen failures, starvation or obvious disease. It primarily occurred in the USA in 2006-07 and was reported from other countries in subsequent years {{3}}.
Comparisons of healthy and CCD-affected colonies showed a correlation between the presence of IAPV and colony collapse, triggering a number of additional studies. In this and a future post I’m going to discuss two of these studies.
I’ll note here that correlation is not the same as causation. Perhaps IAPV was detected because the colony was collapsing due to something else? IAPV wasn’t the only thing that correlated with CCD. It’s likely that CCD was a synergistic consequence of some or all of multiple pathogens, pesticides, poor diet, environmental stress, migratory beekeeping, low genetic diversity and the phase of the moon {{4}}.
IAPV
Israeli Acute Paralysis Virus is an RNA virus. That means the genome is made of ribonucleic acid, a different sort of chemical to the deoxyribonucleic acid (DNA) that comprises the genetic material of the host honey bee, or the beekeeper. The relevance of this will hopefully become clear later.
RNA viruses are not unusual. Deformed wing virus (DWV) is also an RNA virus as is Sacbrood virus and Black Queen Cell Virus. In fact, many of the most problematic viruses (for bees or beekeepers [measles, the common cold, influenza, yellow fever, dengue, ebola]) are RNA viruses.
RNA viruses evolve rapidly. They exhibit a number of features that mean they can evade or subvert the immune responses of the host, they can acquire mutations that help them switch from one host to another and they rapidly evolve resistance to antiviral drugs.
To a virologist they are a fascinating group of viruses.
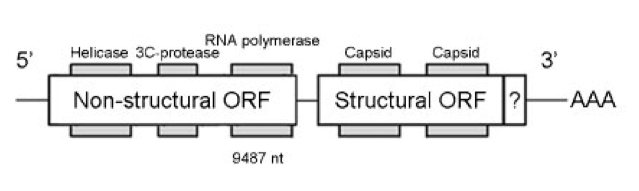
IAPV isn’t a particularly unusual RNA virus. It is a so-called dicistrovirus {{5}} meaning that there are two (di) regions of the genetic material that are expressed (cistrons) as proteins. One region makes the structural proteins that form the virus particle, the other makes the proteins that allow the virus to replicate.
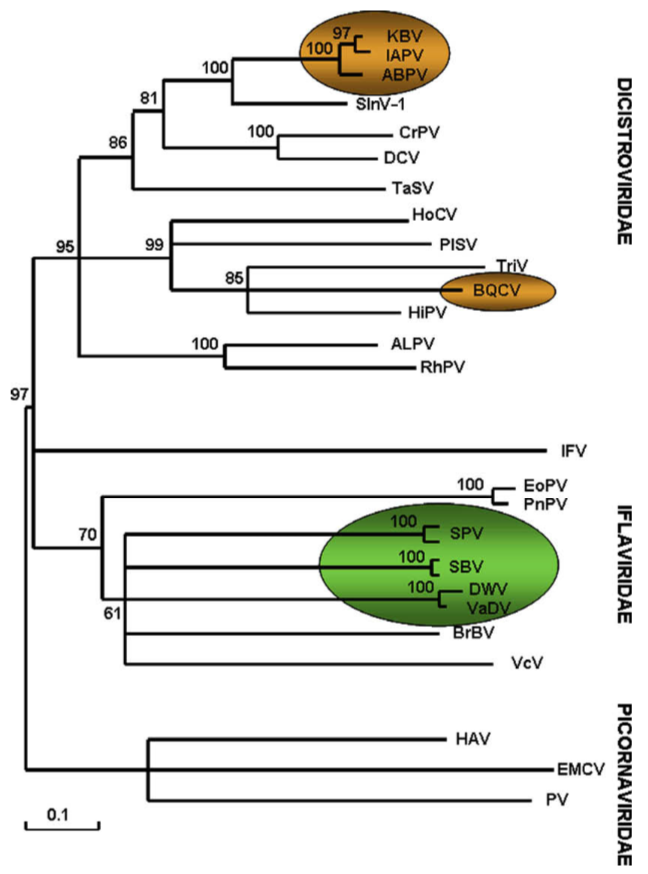
There are many insect dicistroviruses. These include very close relatives of IAPV that infect bees such as Acute Bee Paralysis Virus (ABPV) and Kashmir Bee Virus (KBV). They are very distant relatives of DWV and, in humans, poliovirus; all belong to the picorna-like viruses (pico meaning small, rna meaning, er, RNA i.e. small RNA containing viruses … I warned you about the Latin).
Like DWV, IAPV-infected bees can exhibit symptoms (shivering, paralysis … characteristic of nerve function or neurological impairment in the case of IAPV) or may be asymptomatic. The virus probably usually causes a persistent infection in the honey bee and is transmitted both horizontally and vertically:
- horizontal transmission – between bees via feeding, direct contact or vector mediated by Varroa (not all of these routes have necessarily been confirmed).
- vertical transmission – via eggs or sperm to progeny.
IAPV resistance
An interesting feature of IAPV is that some colonies are reported to be resistant to the virus. This is stated in an interesting paper by Eyal Maori {{6}} but, disappointingly, is not cited.
At the same time these studies were being conducted there was a lot of interest in genetic exchange between pathogens and hosts (e.g. where genetic material from the pathogen gets incorporated into the host) and an increasing awareness of the importance of a process called RNA interference (RNAi) in host resistance to pathogens {{7}}.
Maori and colleagues screened the honey bee genome for the presence of IAPV sequences (i.e. a host-acquired pathogen sequence) using the polymerase chain reaction {{8}}. About 30% of the bees tested contained IAPV sequences derived from the region of the genome that makes the structural proteins of the virus. Other regions of the virus were not detected.
Two additional important observations were made. Firstly, the IAPV sequences appeared to be integrated into a number of location of the DNA of the honey bee (remember IAPV is an RNA virus, so this requires some chemical modifications to be described shortly). Secondly, the IAPV sequences were expressed as RNA. This is significant because RNA is an intermediate in the production of RNAi (with apologies to the biologists who are reading this for the oversimplification and to the non-scientists for some of this gobbledegook. Bear with me.).
And now for the crunch experiment …
Virus challenge
Maori and team injected 300 white eyed honey bee pupae that lacked the integrated IAPV sequence with virus.
Only 2% survived.
They went on to inoculate a further 80 pupae selected at random. Thirteen of these survived (16%) and emerged as healthy-looking adults. The 67 corpses all showed evidence of virus replication and lacked the integrated IAPV sequence in the bee genome.
In contrast, the 13 survivors all contained integrated IAPV sequences but showed no evidence for replication of the virus.
This is of profound importance to our understanding of the resistance of honey bees to pathogens … and in the longer term for the selection or generation of virus-resistant bees.
If it is correct.
Subsequent studies
It’s of such profound importance that it’s extraordinary that there have been no subsequent follow-up papers (at least to my knowledge).
What there have been are number of outstanding but indirectly related studies that have demonstrated a potential mechanism for the integration of RNA sequences into a DNA genome. We also now have a much improved understanding of how such integrated sequences could confer resistance to the host of the pathogen.
Perhaps the best of these follow-up studies is one by Carla Saleh {{9}} on the molecular mechanisms that underlie the integration of viral RNA sequences into the host DNA genome. This study also demonstrates how an acute virus infection of insects is converted to a persistent infection.
One of the big problems with the Maori study is explaining how RNA gets integrated. RNA and DNA are chemically similar but different. You can’t just join one to the other.
Saleh showed the an enzyme called an endogenous reverse transcriptase (an enzyme that converts RNA to DNA) was required. In the fruit fly virus model system she worked with she showed that this enzyme was made by a genetic element within the fruit fly genome (hence endogenous) called a retrotransposon.
Importantly, Saleh also showed that the integrated virus sequences acted as the source for interfering RNAs (RNAi) which then suppressed the replication of the virus.
The study by Saleh and colleagues is extremely elegant and explains much of the earlier work on integration of RNA pathogen sequences into the host genome.

However, it leaves a number of questions unanswered about the bits of IAPV that Maori claim are associated with virus resistance in honey bees.
Unfinished business
The Saleh study is really compelling science. Perhaps the same process operates in honey bees?
This is where issues start to appear. The honey bee genome has now been sequenced. Perplexingly (if the Maori study is correct) it contains few transposons and no active retrotransposons.
Without a source of the reverse transcriptase enzyme there’s no way for the RNA to be converted to DNA and integrated into the host genome.
The second major issue is that there are conflicting reports of the presence of viral sequences integrated in the honey bee genome. The assembled sequence {{10}} appears to contain no virus sequences but there are conference reports of sequences for IAPV, DWV and KBV using a PCR-based method similar to that used by Maori.
Where next?
There’s a lot to like about the Maori study on naturally transgenic bees (a phrase they used in the conclusion to their paper).
It explains the reported IAPV resistance of some bees/colonies (though this needs better documentation). It implicates a molecular mechanism which has subsequently been demonstrated to operate in a number of different insects and host/pathogen systems.
It’s also a result that as a beekeeper and a virologist I’d also like to think offers hope for the future in terms honey bee resistance to the pathogens that can blight our colonies.

Monoculture … beelicious …
However, the absence of some key controls in the Maori study, the lack of any real follow-up papers on their really striking observation and the contradictions with some of the genomic studies on honey bees is a problem.
What’s new?
Eyal Maori has a very recent paper (PDF) on RNAi transmission in honey bees. It was in part prompted by the second of the IAPV studies I want to discuss that arose after IAPV was implicated as a possible cause of CCD. That study, to be covered in a future post, demonstrates field-scale analysis of RNAi-based suppression of IAPV.
It is important for two reasons. It shows a potential route to combat virus infections and, indirectly, it emphasises the importance of continuing to properly control Varroa (and hence virus) levels for the foreseeable future.
{{1}}: Cox-Foster et al., (2007). A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318, 283–287.
{{2}}: And some might make the case that it never existed at all … but that’s a discussion for another time.
{{3}}: There were no well documented or credible cases in the UK, although you will still regularly see articles in the press describing ‘colony collapse’ in the context of UK beekeeping.
{{4}}: OK, not the last one … but the list was extensive and it remains unclear what caused the losses.
{{5}}: Virologists love their Latin!
{{6}}: Maori et al., 2007 Reciprocal sequence exchange between non-retro viruses and hosts leading to the appearance of new host phenotypes. Virology 362:342-3439
{{7}}: Studies had shown that RNAi had a profound range of functions including changes to gene expression and resistance mechanisms in plants and insects. Andrew Fire and Craig Mello shared the Nobel Prize for Medicine and Physiology in 2006 for the discovery of RNAi. Sir David Baulcombe, now of the Sainsbury’s Labs, Cambridge University, should – in my view – have been a co-recipient of this prize.
{{8}}: A sensitive way of detecting genetic material.
{{9}}: Goic et al., (2013) RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nature Immunology 14:396-403.
{{10}}: Honey Bee Genome Sequencing Consortium, (2006). Complete genome sequence and annotation for a social insect, the honey bee Apis mellifera. Nature 443, 931–949.
Join the discussion ...